Note
Go to the end to download the full example code. or to run this example in your browser via Binder
Basic Usage Lesson#
This section demonstrates how to use PyVista to read and plot 3D data using the pyvista.examples.downloads module and external files.
import pyvista as pv
from pyvista import examples
# Set the default plot theme to the "document" theme.
# pv.set_plot_theme('document')
Using Existing Data#
There are two main ways of getting data into PyVista: creating it yourself from scratch or loading the dataset from any one of the compatible file formats. Since we’re just starting out, let’s load a file.
# If you have a dataset handy, like a surface model, point cloud, or VTK file,
# you can use that. If you don't have something immediately available, PyVista
# has a variety of files you can download in its `pyvista.examples.downloads
# <https://docs.pyvista.org/api/examples/_autosummary/pyvista.examples.downloads.html>`_
#
dataset = examples.download_saddle_surface()
dataset
Note how this is a pyvista.PolyData, which is effectively a surface
dataset containing points, lines, and/or faces. We can immediately plot this with:

This is a fairly basic plot. You can change how its plotted by adding
parameters as show_edges=True or changing the color by setting color to
a different value. All of this is described in PyVista’s API documentation in
pyvista.plot(), but for now let’s take a look at another dataset. This
one is a volumetric dataset.
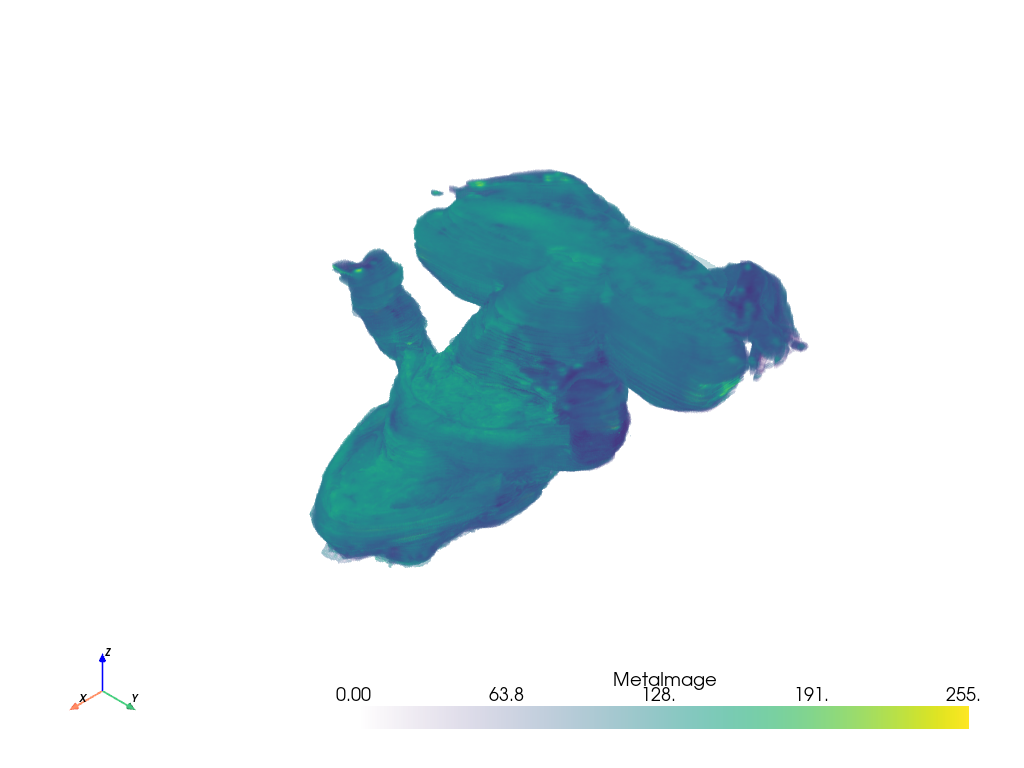
This is a pyvista.ImageData, which is a dataset containing a uniform
set of points with consistent spacing. When we plot this dataset, we have the
option of enabling volumetric plotting, which plots individual cells based on
the content of the data associated with those cells.
dataset.plot(volume=True)
Read from a file#
You can read datasets directly from a file if you have access to it on your environment. This can be one of the many file formats that VTK supports, and many more that it doesn’t as PyVista can rely on libraries like meshio.
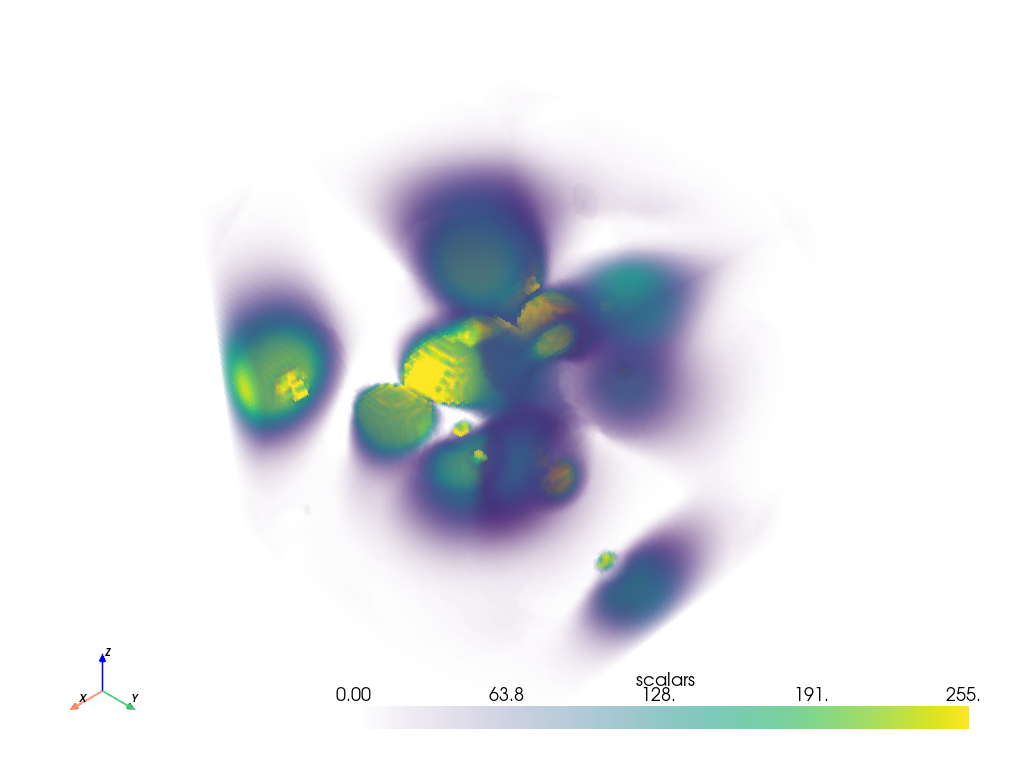
In the following example, we load VTK’s iron protein dataset ironProt.vtk from a
file using pyvista.read().
This is again a pyvista.ImageData and we can plot it volumetrically
with:
dataset.plot(volume=True)
Total running time of the script: (0 minutes 5.899 seconds)
